
Databases for Influenza Virus Research
A Comprehensive Overview of Resources, Tools, and Data Repositories
Key Insights
- Comprehensive Data Integration: Multiple repositories offer integrated genomic, proteomic, and epidemiological influenza data.
- Advanced Analytical Tools: Resources include powerful visualization and analysis tools such as BLAST, phylogenetic analysis, and structural modeling.
- Global Collaboration: International resources facilitate worldwide data sharing, surveillance, and vaccine development efforts.
Overview of Influenza Virus Research Databases
Influenza virus research has significantly benefited from the establishment of numerous databases and resources that facilitate the collection, storage, and analysis of vast amounts of scientific data. These databases provide researchers, epidemiologists, and public health officials with access to genomic sequences, proteomic profiles, structural data, epidemiological trends, and metadata critical for understanding virus evolution, transmission, and pathogenicity. Below is an in-depth discussion of the most notable databases dedicated to influenza virus research.
Influenza Research Database (IRD)
Overview and Features
The Influenza Research Database (IRD) is widely recognized as a cornerstone resource for influenza studies. Sponsored by the U.S. National Institute of Allergy and Infectious Diseases (NIAID), IRD offers an extensive collection of influenza-related data integrated from public repositories, direct submissions, and in-house curation pipelines. Researchers can access genomic sequences, proteomic information, immune epitopes, and epidemiological data through IRD’s intuitive interface.
IRD is equipped with a suite of analytical tools that include BLAST for sequence alignment, multiple sequence alignment programs, phylogenetic tree construction utilities, and 3D protein structure visualization features. The repository’s comprehensive data set is invaluable for hypothesis generation and for designing diagnostics, prophylactics, and therapeutic interventions. With frequent updates and a growing user base, IRD continues to serve as an essential resource in the fight against influenza.
GISAID (Global Initiative on Sharing All Influenza Data)
Overview and Impact
GISAID is one of the most influential databases on a global scale for sharing influenza virus sequence data. It encourages the rapid exchange of viral information during outbreaks and pandemics, thus contributing directly to global public health responses. Unlike many open-access databases, GISAID operates under an access-controlled model that ensures data security while encouraging collaborative research worldwide.
This platform has been critical in supporting epidemiological studies, informing the World Health Organization (WHO) during seasonal vaccine composition recommendations, and tracking mutations and variants across diverse geographic regions. Researchers benefit from GISAID’s extensive metadata documentation and its integration with new analytical models to predict the spread and evolution of influenza viruses.
NCBI Influenza Virus Database
Data Resources and Tools
The National Center for Biotechnology Information (NCBI) offers a specialized Influenza Virus Database that is part of its larger broad-scale genomic repositories. This resource provides a wealth of nucleotide and protein sequence data, accompanied by an array of bioinformatics tools designed for genomic research. The database is incredibly user-friendly, incorporating multiple search and analysis tools that facilitate data mining and comparative genomics investigations.
By redirecting information from the traditional Influenza Virus Resource to the NCBI Virus portal, researchers now have streamlined access to comprehensive viral datasets. These include genome sequences annotated with information on mutations, strain variations, and evolutionary relationships, making it a primary tool for molecular epidemiology research.
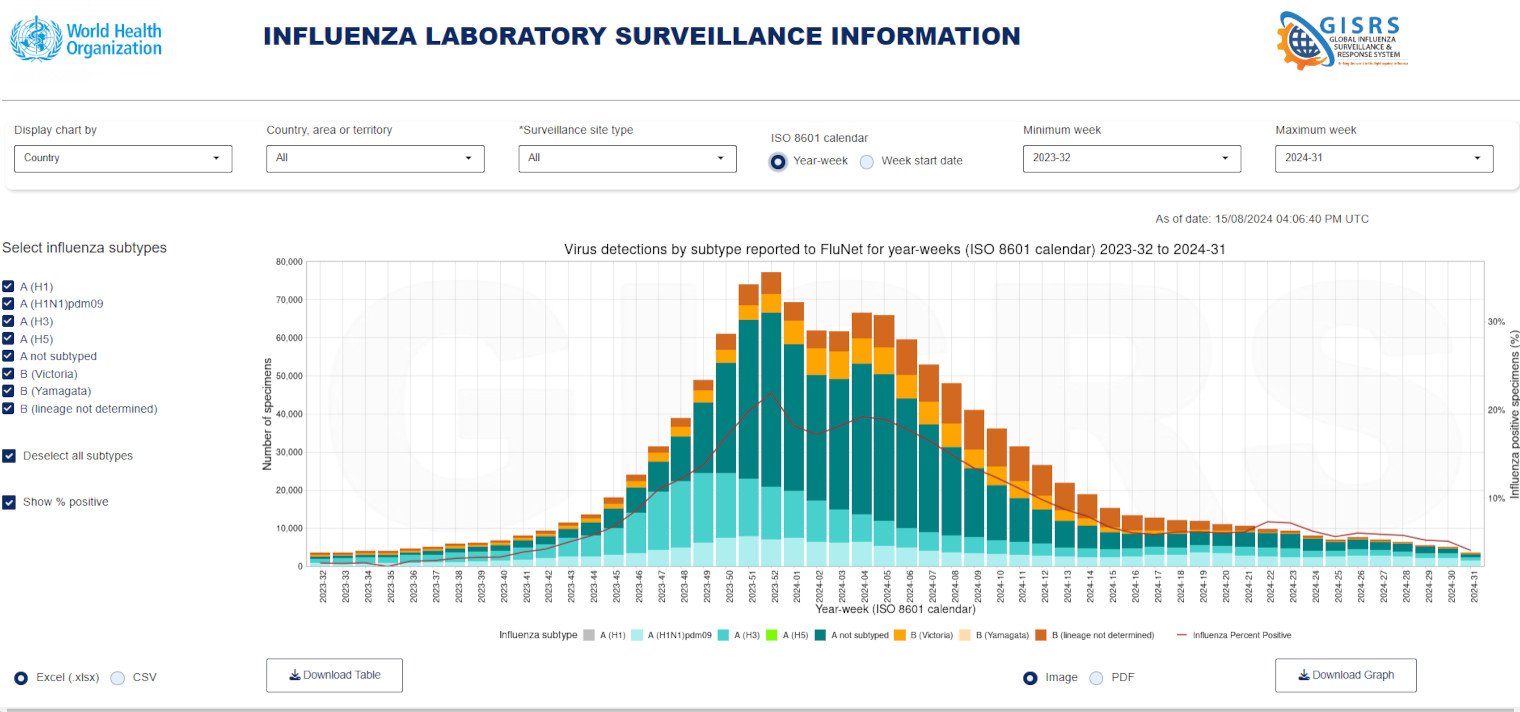
FluNet
Global Surveillance and Epidemiology
Operated by the World Health Organization (WHO), FluNet is a critical tool for global influenza virological surveillance. This web-based tool tracks the occurrence and spread of various influenza strains by aggregating data reported by laboratories worldwide. Public health authorities utilize FluNet for monitoring trends in influenza activity, which in turn informs responses to outbreaks and seasonal epidemics.
The regular updates provided by FluNet enable timely decision-making regarding public health interventions, vaccine formulation, and resource allocation during significant influenza outbreaks. Its interface is designed to provide both an overview of global influenza trends and detailed data on subtype distributions.
Additional Databases and Resources
Nextflu, ViPR, and Others
Beyond the primary databases, several additional resources offer complementary data for influenza research:
- Nextflu: Nextflu provides real-time tracking of influenza virus evolution by analyzing genetic sequence data. It allows researchers to model the evolution and predict future changes in the virus, making it a valuable tool for anticipating future strains.
- Virus Pathogen Database and Analysis Resource (ViPR): ViPR supports the study of viral pathogens, including influenza, by providing tools for sequence analysis, visualization, and comparative genomics. The platform aggregates data from multiple sources, enabling researchers to identify patterns in viral evolution.
- OpenFluDB: A user-driven database that allows for community engagement and data submission, OpenFluDB is an evolving resource aimed at expanding transparency and access to influenza virus data.
- Bacterial and Viral Bioinformatics Resource Center (BV-BRC): Although this resource focuses on a broader range of pathogenic organisms, its dedicated influenza modules offer detailed genetic and proteomic information that supports integrative research approaches.
Each of these secondary platforms adds value by offering specialized tools or data sets that enhance our understanding of influenza viruses from different perspectives, ranging from evolutionary biology to public health surveillance.
Comparative Overview of Key Databases
To further illustrate the differences and similarities among these resources, the following table summarizes the key features, primary uses, and access modalities of the most frequently used influenza databases.
| Database | Primary Data | Key Features | Access |
|---|---|---|---|
| Influenza Research Database (IRD) | Genomic, proteomic, immune epitopes, surveillance data | Analysis tools (BLAST, phylogenetics, visualization) and integrated data pipelines | Open-access |
| GISAID | Viral genome sequences, epidemiological metadata | Global data sharing, secure data access, contribution to vaccine composition | Registration required |
| NCBI Influenza Virus Database | Nucleotide and protein sequences with rich metadata | Robust search, comparative genomics, integration into larger NCBI resources | Open-access |
| FluNet | Laboratory surveillance data on influenza virus subtypes | Real-time global surveillance, trend analysis, public health tool | Open-access |
| Nextflu | Genetic sequence data | Real-time evolution tracking, predictive models | Open-access |
Details on Data Collection and Usage
The value of these databases extends beyond simply providing raw data. They serve a myriad of purposes in the influenza research community:
Data Integration and Curation
Many of these resources, such as IRD and NCBI, employ rigorous data curation protocols that integrate information from numerous sources. This integration ensures that data is standardized, annotated, and presented in a format that facilitates meaningful biological insights. For instance, the IRD aggregates data from GenBank, direct laboratory submissions, and internal curation efforts to provide a consistent platform for comparative analysis.
Advanced Analytical Tools
Integrated analytical tools are a hallmark of these databases. Researchers can:
- Use sequence alignment tools to identify similarities and differences among strains, which can reveal patterns of mutation and recombination.
- Construct phylogenetic trees to trace the evolutionary history of influenza viruses. This is indispensable when studying antigenic drift and shift events.
- Visualize protein structures and epitope mapping to understand how immune pressures shape viral evolution.
In addition, these tools often allow for the integration of epidemiological data, linking genomic changes with real-world disease prevalence and geographic spread. This synergy between data types accelerates the understanding of viral behavior, which is crucial for the development of effective vaccines and therapeutics.
Global Collaboration and Data Sharing
One of the most significant advancements in the field is the facilitation of global collaboration. Resources like GISAID and FluNet empower scientists and public health officials around the world to share data rapidly and securely. This not only enhances global preparedness during influenza outbreaks but also supports ongoing research into virus evolution and interspecies transmission.
In crisis situations, such as the emergence of new pandemic strains, these platforms allow for real-time data sharing and analysis, thereby informing swift public health responses and guiding vaccine development in a timely manner.
Comparative Features of Data Portals
To further enhance understanding of these portals, a more detailed comparative table is presented below:
| Aspect | IRD | GISAID | NCBI Influenza Database | FluNet |
|---|---|---|---|---|
| Data Types | Genomic, proteomic, immune epitopes | Sequence and epidemiological metadata | Nucleotide, protein sequences, annotations | Subtype prevalence, surveillance reports |
| Analytical Tools | BLAST, phylogenetic trees, visualization tools | Data sharing, mutation tracking | Search and retrieval, comparative genomics | Real-time monitoring, trend analysis |
| Access Policy | Open-access | Access-controlled (registration) | Open-access | Open-access |
Utilizing Influenza Databases in Research
Practical Applications
Researchers can utilize these databases across a range of applications to advance the understanding of influenza viruses. For example, during periods of seasonal outbreaks or emerging pandemics, real-time genomic sequencing data combined with epidemiological surveillance data helps in:
- Epidemiological Tracking: Monitoring spread patterns and identifying the introduction of new variants in different geographic regions.
- Vaccine Development: Analyzing trends in antigenic drift and shift to make informed decisions on vaccine strain selection.
- Drug Target Identification: Investigating structural data to identify potential targets for antiviral drugs and therapeutic interventions.
- Molecular Evolution Studies: Utilizing phylogenetic analysis tools offered by these databases to understand the evolutionary relationships and mutation patterns among influenza strains.
By combining diverse data types and advanced analytical tools, these repositories not only facilitate immediate and practical research needs but also lay the foundation for long-term studies that can lead to breakthrough insights into viral behavior and pathogenesis.
Integration with Multi-disciplinary Research
Modern influenza research is notably interdisciplinary, integrating bioinformatics, molecular biology, immunology, and epidemiology. The databases mentioned above provide a seamless interface where data from these varied disciplines can be cross-referenced. For instance, researchers might integrate genetic sequence data from the NCBI Influenza Virus Database with epidemiological insights from FluNet to model outbreak dynamics, or they may combine immunological data from IRD with computational analysis from ViPR to predict antibody binding sites on viral proteins.
Such multi-faceted approaches enrich our understanding of virus-host interactions and open new avenues for targeted therapeutic interventions. The collaborative nature of these databases fosters an environment of open scientific inquiry, wherein researchers share methodologies, compare findings, and build upon each other’s work, thus accelerating the pace of discovery.
Final Thoughts on the Role of Influenza Databases
The establishment and continuous improvement of influenza virus research databases represent a quantum leap in our ability to monitor, study, and ultimately control influenza outbreaks. Through the integration of global data sharing, sophisticated bioinformatics tools, and interactive analytical platforms, these databases have transformed the approach to influenza research from isolated efforts to a comprehensive, interconnected global network. This advancement is pivotal for both routine public health monitoring and rapid responses to emerging influenza strains.
Researchers and public health professionals continue to rely heavily on these resources, as evidenced by their widespread citation in over hundreds of scientific publications and their routine use in clinical and laboratory settings. As the field of influenza research evolves—with advancements in genomic sequencing technology and data analytics—the integral role played by these databases will become even more apparent, underscoring their importance in antibiotic research and public health preparedness.
References
- Influenza Research Database - Wikipedia
- NCBI Virus Database - NCBI
- GISAID - Global Initiative on Sharing All Influenza Data
- FluNet - WHO
- Influenza Research Database (IRD) - J. Craig Venter Institute
Recommended Related Queries
Last updated March 11, 2025